ACCELERATOR NEWS
Progress of dOPM usage from Edinburgh
Accelerator team members from the CRUK Scotland Centre based at the Institute of Genetics and Cancer, University of Edinburgh are working on translating the dOPM microscope system for use in high content assays.
The initial work is focussing on assessing the reproducibility of experiments in a spheroid model of glioblastoma using NPE cells.
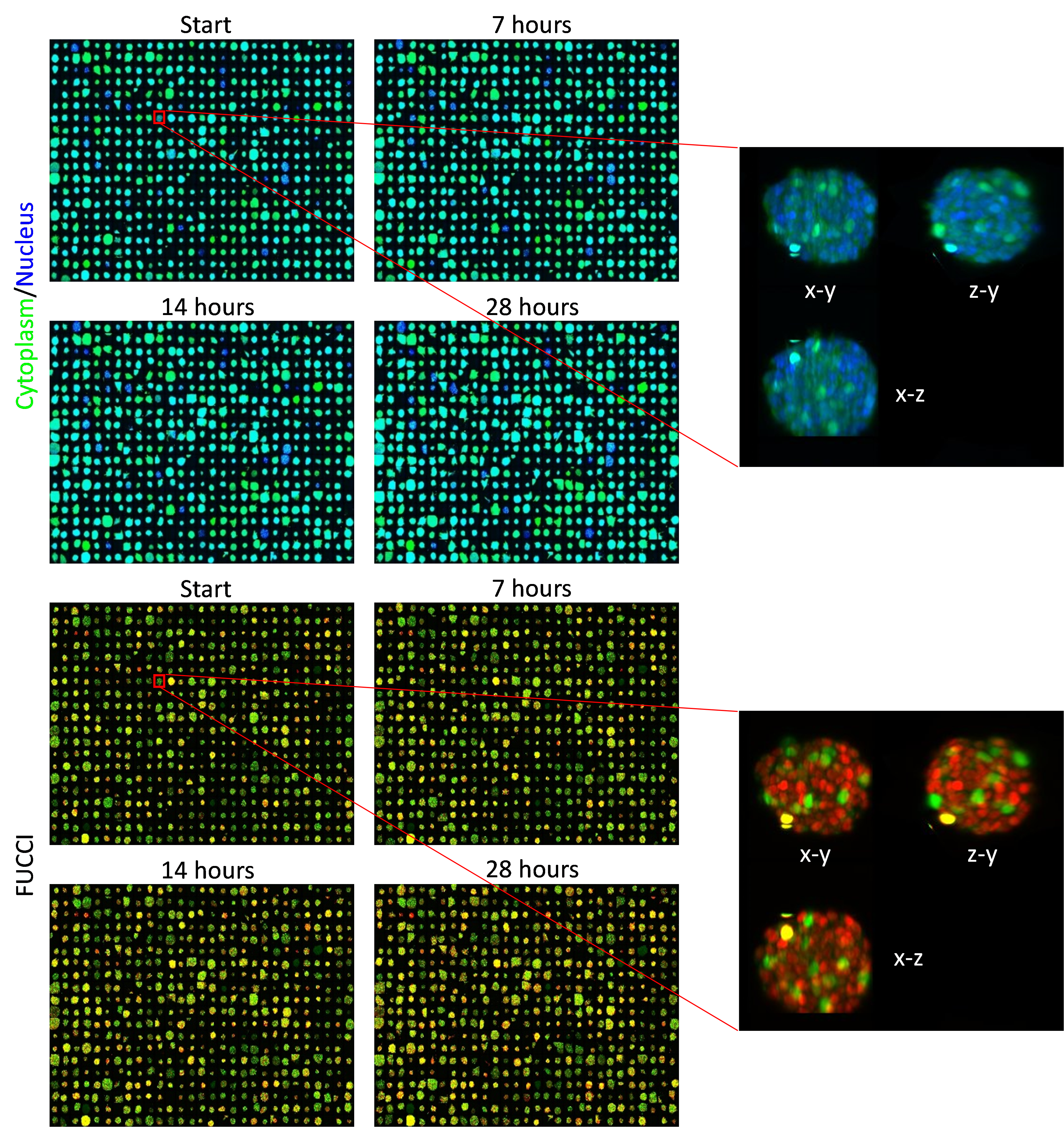
In high content screening, the samples to be imaged are arrayed in multi-well plates. In the image above, we have performed 3D imaging of ten spheroids per well, in fifty-eight wells and this has been repeated across four time points as the spheroids grow. Each image shows a montage of all five hundred and eighty spheroids. The top four panels in green/blue show the cytoplasm/nucleus respectively at each of the four timepoints, and the bottom four panels show the readout of the FUCCI cell-cycle reporter for the same spheroids; the FUCCI reporter changes the colour of its emission through red to yellow to green as the cell progresses through its division cycle. The cell-cycle information can also be used to measure the effects different drug treatments will have on the cells. You can also see the cells getting slightly bigger with each passing time point.
This experiment is being repeated at least three times, which will allow us to accurately measure the variation in signal between experiments. This is an important step in producing reliable drug screening assays, as it allows us to know when a drug is affecting spheroid growth.
The team are also using the dOPM system to measure these changes continuously over time. In the video, we can see the effect of the drug staurosporine that is known to kill cells. We can see the spheroids are destroyed by the drug at both concentrations, whereas our control spheroids treated with the non-toxic DMSO remain unaffected.
Size of scale bar: 50µm.
Labels: Cytoplasm, FUCCI G1, FUCCI G2, FUCCI S
Image Technologists awarded Provost Award for Laser Safety

Sunil Kumar and Hugh Sparks, our Imperial-based Accelerator technologists who also work in our satellite laboratory at the Crick, have been awarded the 2023 Provost’s award for Excellence in Safety at Imperial College London. This recognised their work developing practical laser safety training and an online risk assessment resource for developers and users of our advanced microscope systems.
MACH3CANCER on the Road

Paul French gave an invited talk at the Society for Laboratory Automation and Screening (SLAS) annual meeting 2023, San Diego, USA on ‘Implementing advanced microscopy modalities to increase content and throughput – while widening access’.
Chris Dunsby gave an invited talk at the NOTICE Glasgow symposium on ‘High-speed 3D light-sheet microscopy’
Accelerator PhD students Billy Flanagan, Huihui Liu and staff member Liuba Dvinskikh attended and presented at Focus on Microscopy (FOM) 2023 in Porto, Portugal in April. The conference brings together develops and users of optical microscopy and provides a platform for the dissemination of information about new developments. They presented talks on ‘Single-shot semi-quantitative phase contrast microscopy’ and ‘Open-source multibeam multiphoton multiwell plate microscope’ and a poster on ‘Dual-view oblique plane microscopy for high-content imaging of complex and heterogeneous 3D cancer organoid models’.
Accelerator technologist, Hugh Grant, gave an invited talk at the SLAS 2023 Building Biology in 3D Symposium in Cambridge on ‘High speed and high content light sheet fluorescence microscopy’ and also gave a talk titled ‘Dual-View Oblique Plane Microscopy for Multi-Well Plate Time-Lapse 3D Imaging Assays in Cancer Research’ at the Optica Biophotonics Congress 2023 Conference in Vancouver.
OPM implemented at the Institute of Cancer Research, London
A new dOPM system has been implemented at the Institute of Cancer Research in Chelsea, London by Liuba Dvinskikh under the guidance of Hugh Sparks and Chris Dunsby. A launch event at the institute was held on Feb 20th 2023, featuring talks on the Mach3Cancer project and OPM, as well as a demo of the system. The OPM system is being applied to imaging 3D cancer models developed by the Chris Bakal group.
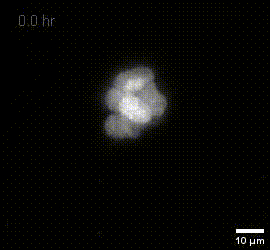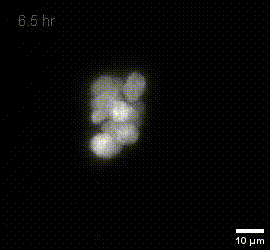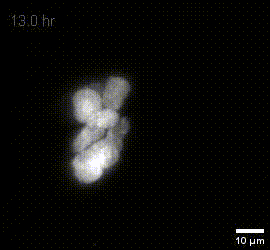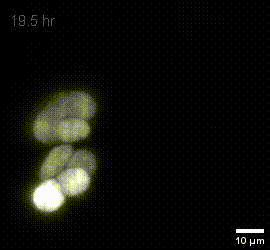
MIP of fused dOPM timelapse data over 19.5 hours at 30 minute intervals of live U2OS Spheroids with H2B mCherry prepared by Vicky Bousgouni and imaged by Liuba Dvinskikh.

Image stack flythrough through processed single views 1 & 2 and fused data of fixed melanoma spheroids with fixed melanoma spheroids with EGFP-CAAX and histone-H2B prepared by Lucas Dent and imaged by Liuba Dvinskikh.
OPM used by CRUK Edinburgh Centre to image 3D glioma mass
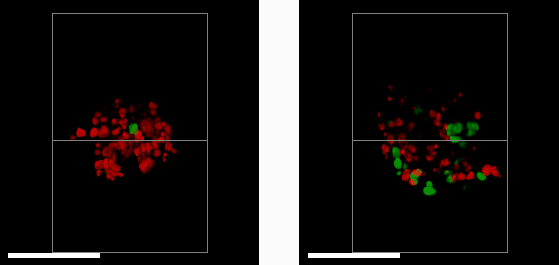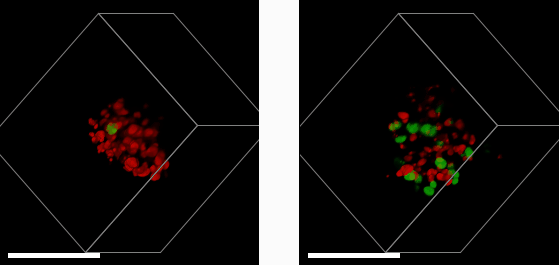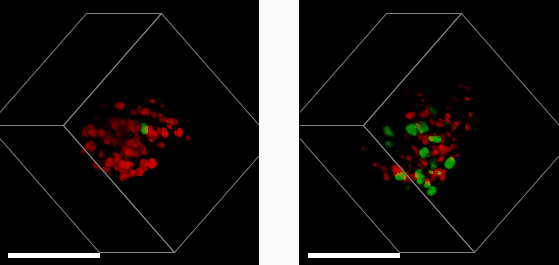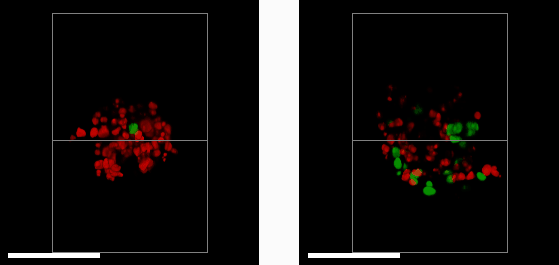
E57 Mesenchymal Glioma with Fucci reporter (Sartorius 4779) treated with 1μM PF05212384 (left) or DMSO control (right) imaged after 3 days treatment.
Researchers at the Edinburgh CRUK centre have acquired their first image data using their new oblique plane microscope (OPM) system
A 3D mass of glioma (brain tumour) cells derived from a patient with a mesenchymal sub-type (the most aggressive glioma subtype) was treated with a drug that halts cell growth and division (left). This is measured using a reporter system that colours cell nuclei that are ready to divide and grow in green, and those that aren’t in red. By comparing to a control sample (right), we see that the drug (left) has greatly reduced the number of dividing cells. Studying cells in a 3D environment maximises cell-cell and cell-environment contacts as well as causes gradients of nutrients, oxygen and therapeutic drug to form across the 3D structure. This more closely resembles the behaviour of tumours in their native environment – compared to cell culture on in a monolayer of cells – providing a more accurate tool to help study cancer treatments.
OPM implemented at the CRUK Edinburgh Centre

Hugh Sparks and Nils Gustafsson implemented a new dOPM instrument at the CRUK Edinburgh Centre in the phenotypic discovery facility at the University of Edinburgh, under the guidance of Chris Dunsby. This is being applied to image cancer organoids and other 3D cancer models by Jayne Culley and Martin Lee under the guidance of Neil Carragher.
Progress on OPM implemntation at IRB Barcelona
Nils Gustafsson implemented a new dOPM instrument at the IRB in Barcelona under the guidance of Chris Dunsby. This system is being integrated into the Advanced Digital Microscopy facility led by Julien Colombelli and will be exploited by the lab of Eduard Batlle.
OPM implemented at the Francis Crick Institute

The first CRUK Accelerator OPM instrument has been implemented and upgraded at the Francis Crick Institute
Nathan Curry and Nils Gustafsson implemented a new OPM instrument at the Francis Crick Institute under the guidance of Chris Dunsby. This was applied to image cancer organoids and has now been upgraded to a new design with higher resolution and signal to noise performance. This new OPM design will now be implemented for our other Accelerator partners at the Institute of Cancer Research, the University of Edinburgh and IRB Barcelona.
OPM image data– acquired at Imperial
Oblique Plane Microscopy (OPM) is a core technology for our Accelerator that we will implement at our partner sites. Here we present renderings of exemplar OPM image data of 3D cancer models from our consortium partners that have been imaewd on the first prototype OPM instrument at Imperial.
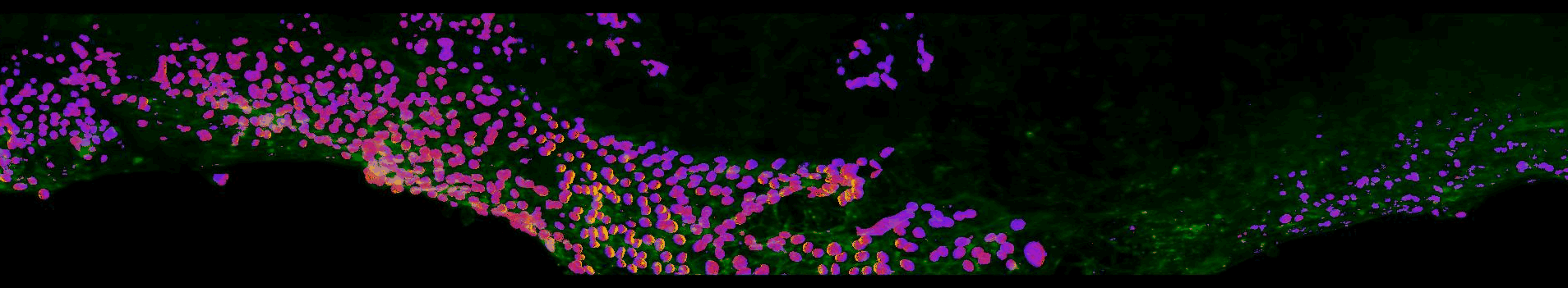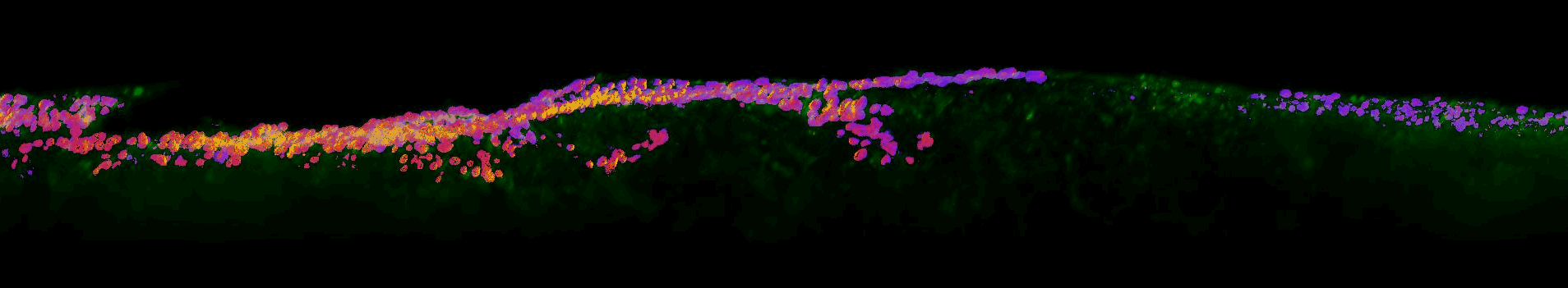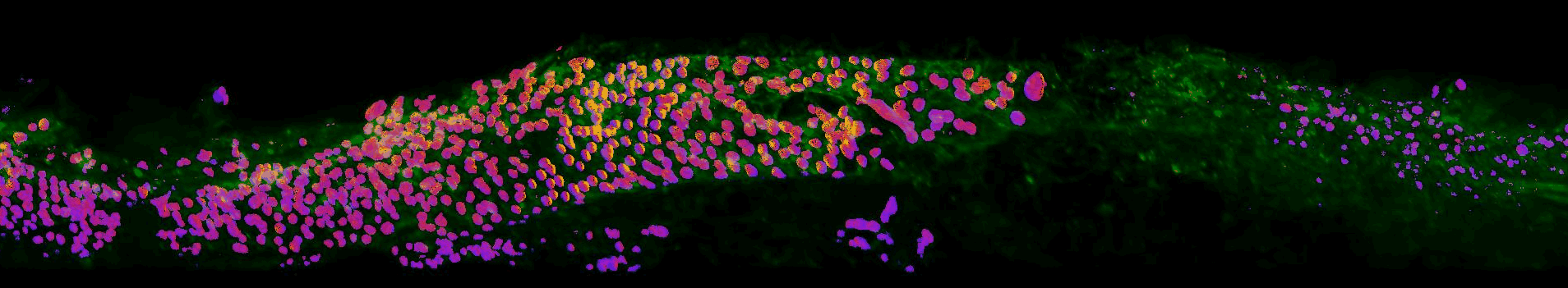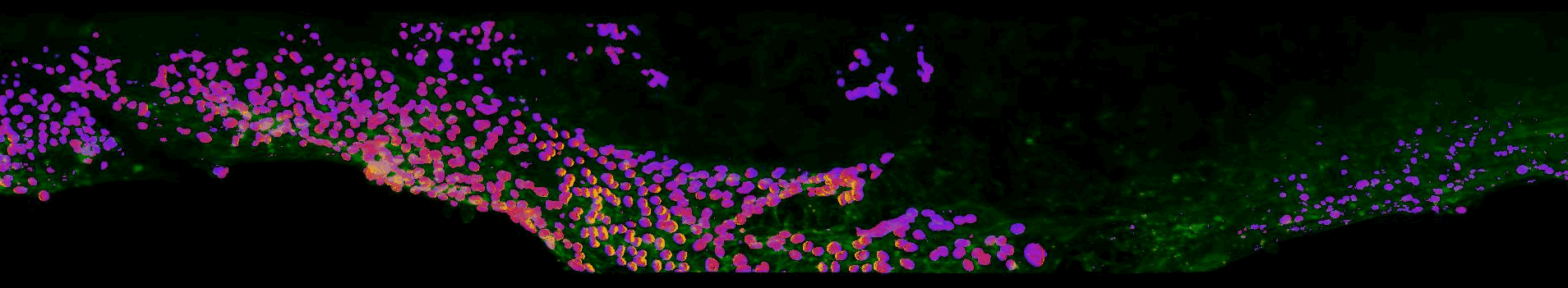
Sections of EGFR mutant mouse lung tissue treated with latest EGFR inhibitors- Sahai lab, Crick
This work aims to improve our understanding of residual disease and its microenvironment after treatment. The figure above shows a section of mouse lung tissue treated with an EGFR inhibitor imaged using the OPM system. Actin is labelled in green. The yellow-red-purple false-colour indicates the FRET ratio reported by the YPet/ECFP EKAR-EV FRET biosensor for ERK activity. The field of view shown is approximately 1.5×0.32×0.14 mm3.
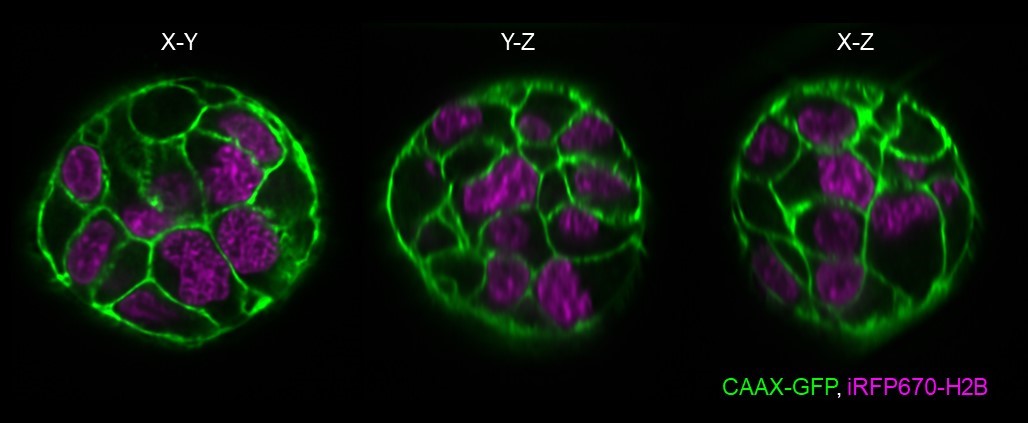
Melanoma spheroids – Bakal lab, ICR
We are interested in how changes in cell size and shape drive melanoma metastasis in 3D. We aim to study this using melanoma spheroids expressing mutant NRAS (G12D) and also expressing membrane (CAAX-GFP) and nuclear (iRFP670-H2B) labels. The spheroids were cultured in reduced growth factor basement membrane extract (BME). Fifteen spheroids were imaged in the same experiment at 15 min intervals. The dOPM images above show a near-isotropic resolution in all three dimensions, which will aid the automated extraction of cell shape features for analysis.
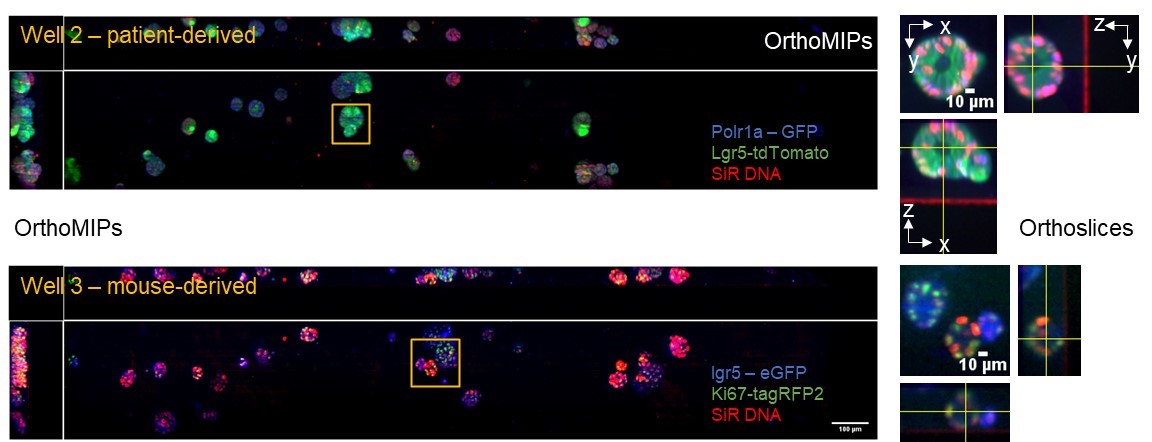
Patient and mouse-derived colon cancer organoids – Battle lab, IRB
We aim to quantify the dynamics of cancer stem cell division and differentiation using engineered PDOs where specific marker genes are labelled to quantify the proportion of proliferating and non-proliferating stem cells. Readouts in this work include the specific markers for stem cells, LGR5-GFP and Ki67-RFP. We imaged 8 fields in a multi-well plate where the field of view was 2×0.32×0.14 mm3 and the image acquisition time was 3 minutes per field for 3 colours. The figure above shows representative data from patient-derived (well 2) and mouse-derived (well 3) organoids acquired using the OPM system .
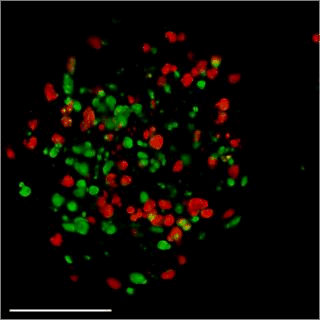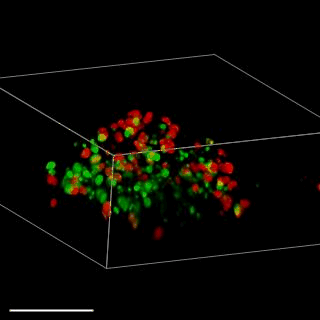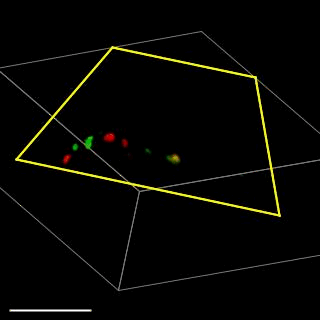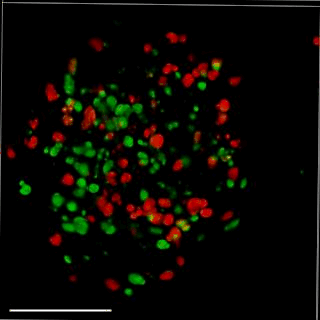
Patient-derived glioma stem cell spheroid model – Carragher lab, CRUK Edinburgh
We are aiming to develop an automated 3D phenotypic screening assay to identify small therapeutic molecules targeting the quiescent glioma stem cell niche near the hypoxic core and not the proliferating glioma stem cells on the periphery of the spheroid. The figure above shows a fixed patient-derived glioma stem cell spheroid model expressing the FUCCI cell cycle reporter imaged using OPM.
GFP (green) nucleus – G1 phase cell cycle
Cherry (red) nucleus – S phase cell cycle
Red and green – G2/ML phase cell cycle