CONTEXT
Resistance to cancer therapy arises from heterogeneous cellular responses within tumours grown in complex 3D environments but this cell-to-cell variability is rarely addressed in standard in vitro drug screens, which typically measure the average response of cell populations in highly artificial contexts, often confusing real biological variance with noise and failing to capture the behaviour of outliers that may drive drug resistance. While some progress has been made to study phenotypic heterogeneity by single cell-resolved assays of 2D cell cultures using current high content analysis (HCA) technologies, the failure to account for heterogeneous therapeutic responses in complex 3D tumour environments, including interactions with other cells such as fibroblasts and immune cells, contributes to the inefficiency of cancer drug discovery programmes and poor outcomes for many cancer patients.
Developing new cancer therapies requires improved understanding of heterogeneity in cell behaviour. Assays for new potential cancer therapies must therefore address single-cell responses, rather than the average responses of cell populations that fail to account for outlier cells that can be responsible for drug resistance. This will require single-cell-resolved imaging capabilities that can be employed at reasonable throughput. Since individual cancer cell behaviour will depend on the local tumour microenvironment (TME), it is also necessary to progress from assays of homogeneous monolayers of cells (typically using immortal cell lines) to more complex 3D cancer models, such as patient-derived organoids (PDO), that may better recapitulate the complexity of the in vivo context. However, increasing the physiological complexity of cancer models also increases their opacity and scale – limiting opportunities for high throughput (phenotypic) screening. Current high throughput assays involving these complex 3D models are typically limited to relatively simple endpoints since the HCA instrumentation and expertise to image diverse responses with single cell resolution in extended 3D cell cultures are not yet commercially available. Furthermore, the reproducibility of (heterogeneous) responses to drugs in 3D cancer models, and therefore the reliability of such assays, are not yet established.
This project aims to accelerate the development and deployment of 3D HCA capabilities to enable single-cell resolved assays of complex 3D cancer models to enhance cancer drug discovery.
STRATEGY
We will develop and share modular open source automated high content analysis (HCA) instrumentation adapted for 3D imaging of complex cell cultures presenting a range of optical properties.
We will also explore cell culture and sample preparation (e.g. labelling, mounting, clearing) techniques in order to help cancer researchers optimise the trade-off between the complexity of cancer models and the power of assays in terms of (single cell) resolution and throughput.
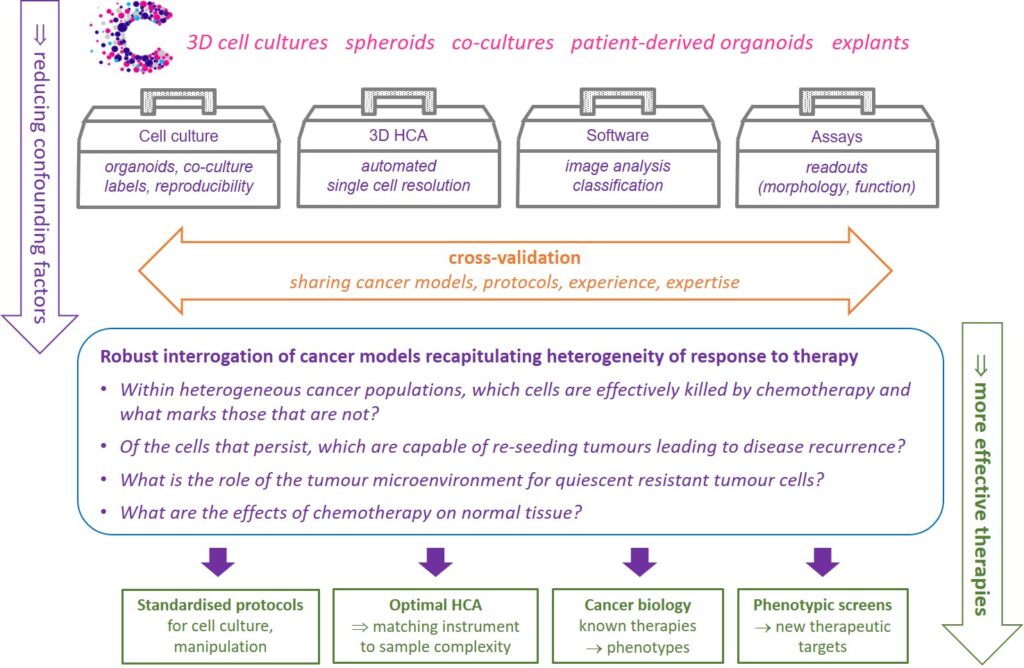
We will demonstrate these new Accelerator capabilities through assays addressing exemplar cancer biology questions, determining which cells within a heterogeneous cancer population are effectively killed by chemotherapy and which persisting cells drive disease recurrence.
We will also explore the role of the tumour microenvironment for quiescent resistant tumour cell sub-populations. To better understand side-effects of chemotherapy, we will assay its impact on the regenerative capacity of stem cell models of normal tissue.
The new Accelerator infrastructure, know-how and knowledge gained will enhance cancer biology and drug discovery, informing the development of new assays and commercial instrumentation.
TOOLS AND TECHNOLOGIES
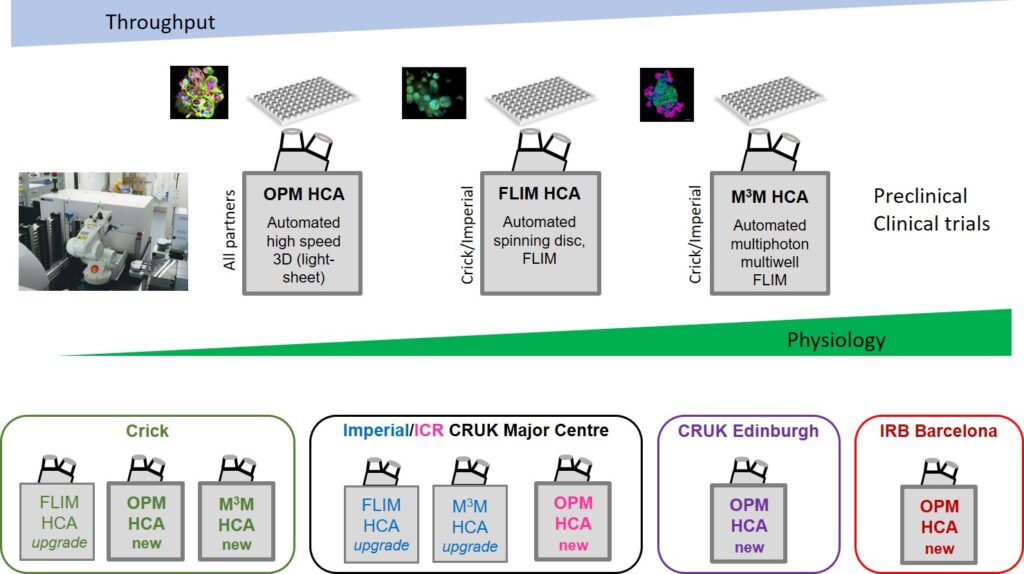
We will develop a suite of modular, open source automated microscopes providing 3D imaging of organoids and other samples arrayed in multiwell plates. These HCA capabilities will include spinning disc confocal microscopy systems adapted for FLIM for robust automated single cell-resolved (FRET) readouts of cell signalling and drug-target engagement in fixed and live PDO. For higher throughput, we will implement automated light-sheet multiwell plate imaging using oblique plane microscopy (OPM) to provide rapid (>25 volumes/s) 3D imaging of cell morphology and dynamics up to ~100 μm depth. For larger and/or more scattering PDO, we will implement adjustable multibeam, multiphoton microscopy (MMM) with adaptive optics to provide automated 3D imaging and FLIM (including label-free readouts of metabolism using cellular autofluorescence). These capabilities will be developed and implemented in our photonics laboratories at the ICR/Imperial CRUK Major Centre (in the Physics Department and at the ICR Chester Beatty labs) and at the Crick. Of these technologies, OPM is potentially the most immediately impactful advance in 3D HCA and this capability will also be implemented in our partner laboratories at CRUK Edinburgh and IRB Barcelona. Across the consortium, we will develop bespoke open source software for 3D image analysis, and protocols and SOPs for preparing and imaging PDO. Standardized assays will be developed for cross-validation between Accelerator partners.